Our laboratory is focused on understanding the allosteric mechanisms that allow macromolecular complexes to perform and regulate their function.
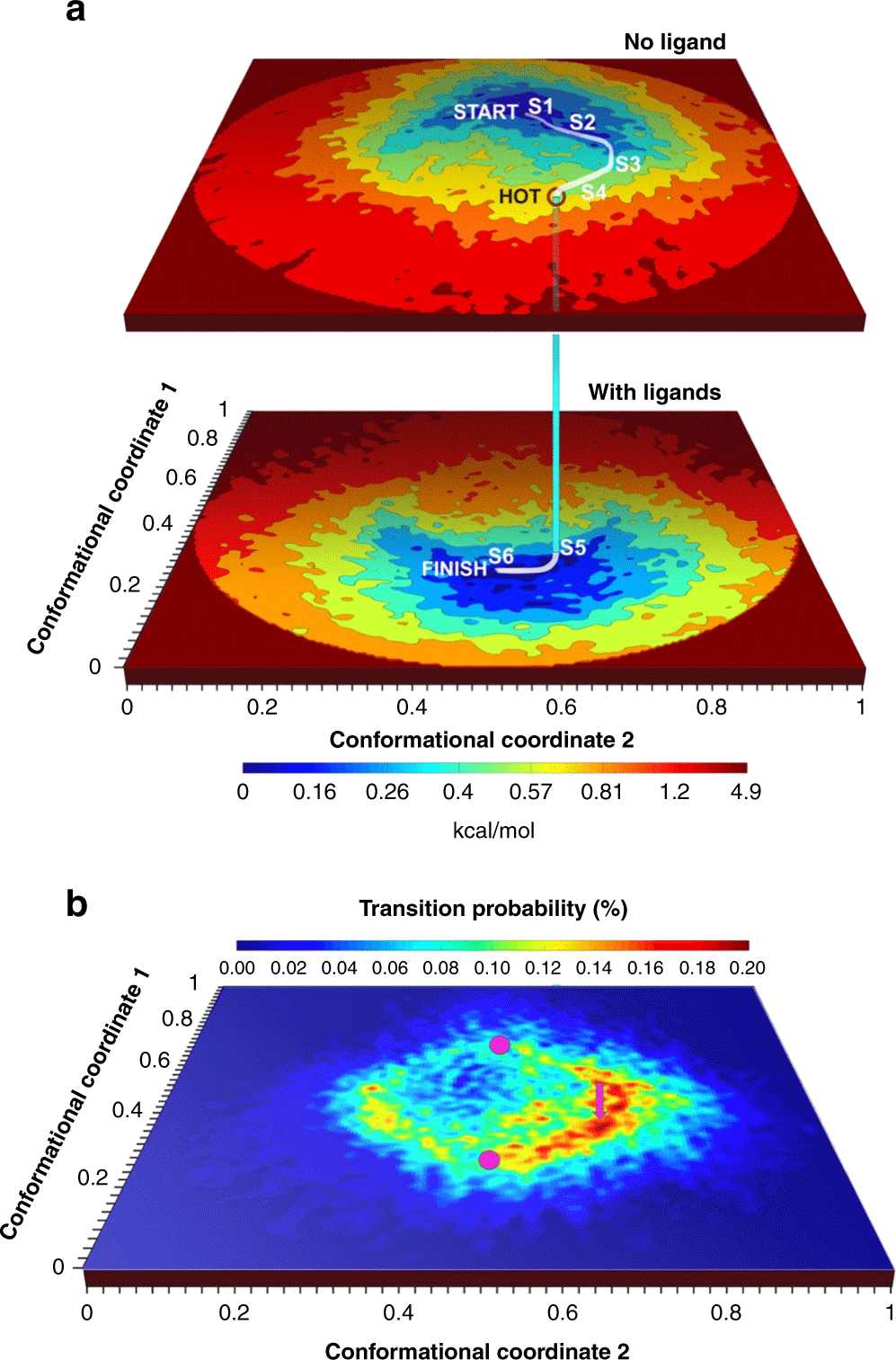
Cryo-electron microscopy gives us the unique opportunity to obtain experimental molecular movies to depict these macromolecules in action, a paramount goal of structural biology. We aim to use this unique opportunity to shed light on pharmacologically-relevant processes with a particular focus on mechanisms happening at the cell membrane.
Signaling at the Membrane
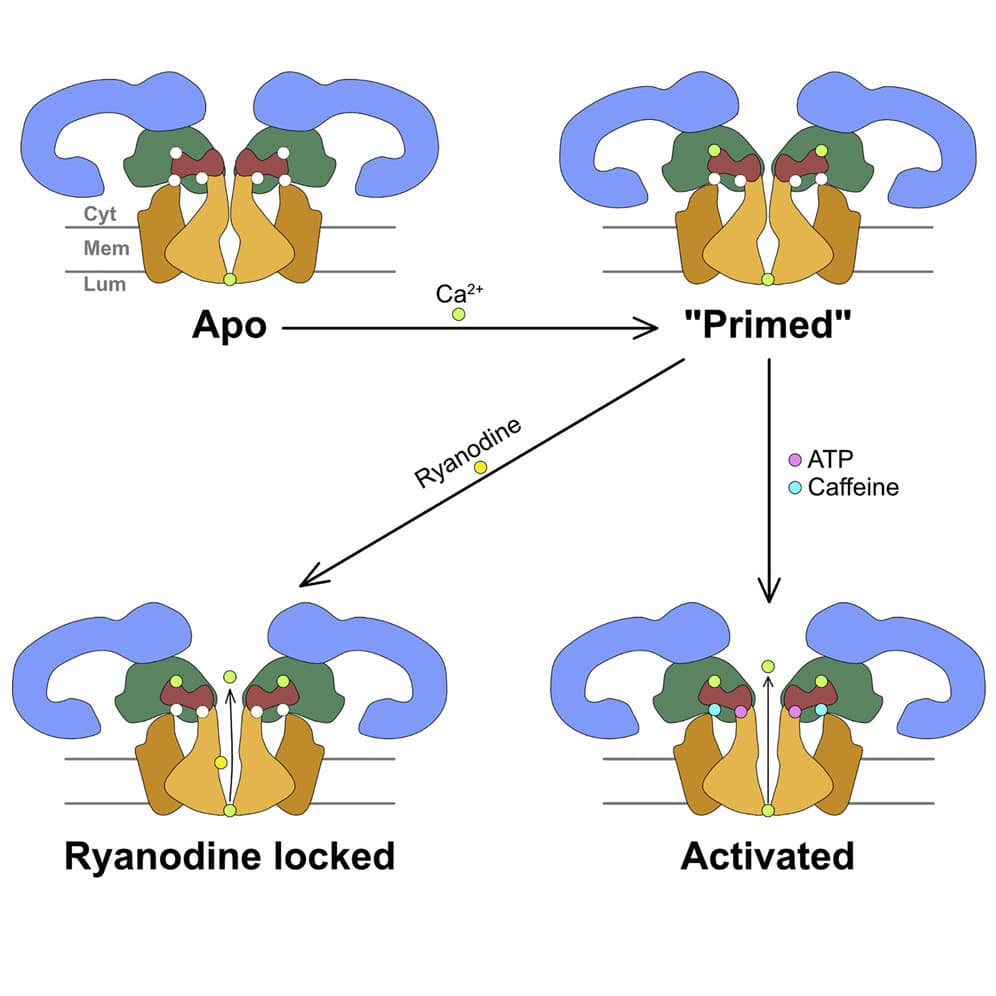
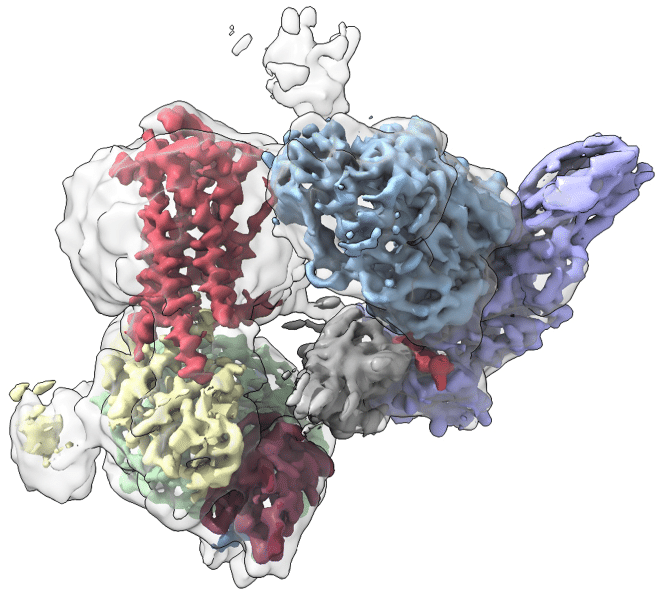
Ryanodine receptors
G protein-coupled receptors
G protein-coupled receptors (GPCRs), also alternatively referred to as seven transmembrane receptors (7TMRs), are the largest class of cell surface receptors and are involved in the regulation of many physiological processes. Their ubiquity in physiology and disease as well as their diversity in structure and function make them very attractive targets for therapeutic development. Currently, it is estimated that over one third of all approved therapeutics target GPCRs.
We aim to take advantage of this golden age that cryo-electron microscopy is bringing to the structural study of GPCRs. Cryo-electron microscopy allows us to study ligand states previously inaccessible and to peer into their dynamics, directly retrieved from the electron microscopy data. This will be particularly useful for structure-based drug discovery efforts and for the development of receptor subtype-specific drugs or biased ligands for the treatment of a variety of diseases.
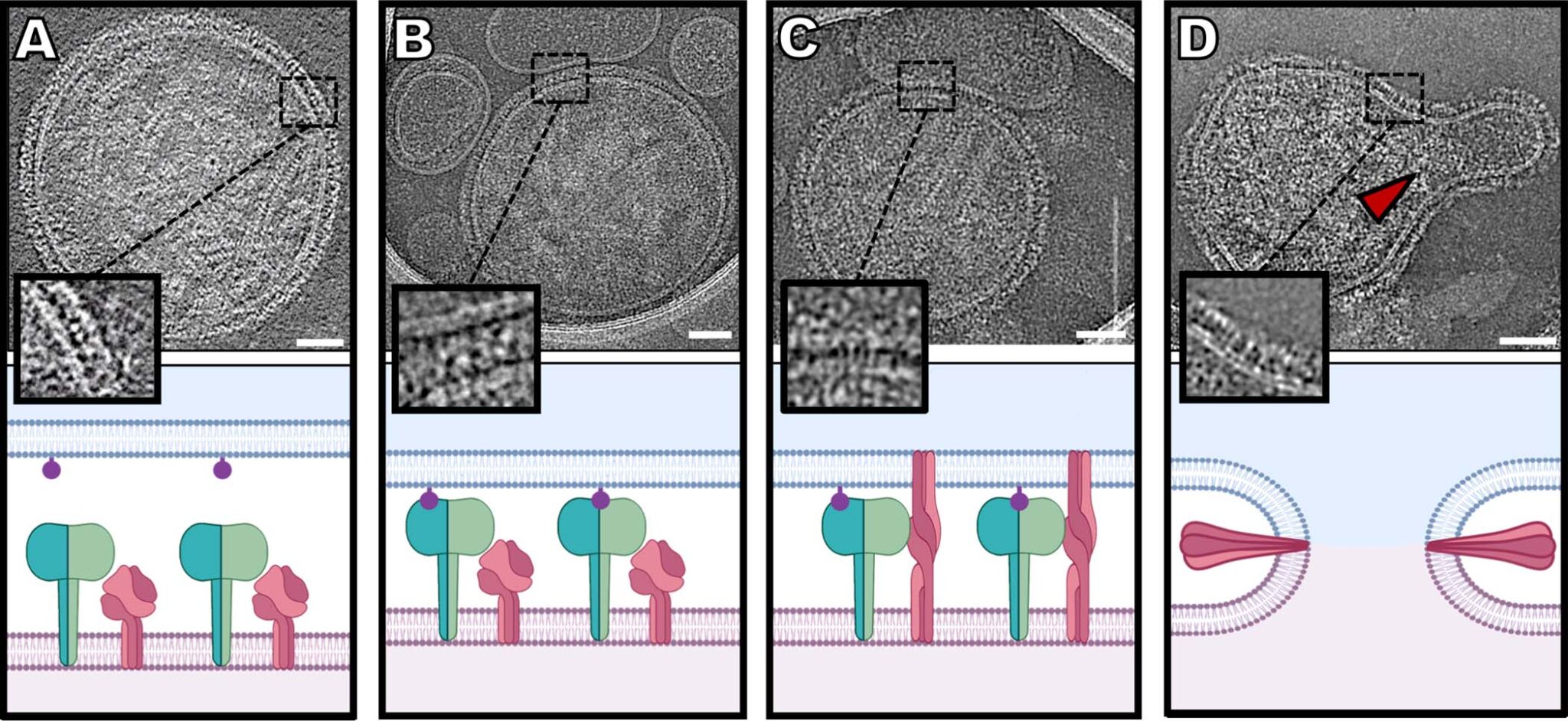
Host-Pathogen interactions
The recent pandemics, including the current COVID-19 pandemic, are showing us how global warming and other changes in our environment are facilitating the transfer of new diseases to human populations, with devastating consequences. At the heart of this problem is the adaptation of pathogens to interact with new hosts. Uncovering the molecular mechanisms underlying these host-pathogen interactions are integral in developing new strategies to prevent or cure infectious diseases. We aim to uncover the dynamic molecular events from initial interaction to entry of pathogens into their host with a combination of single-particle and tomographic cryo-electron microscopy. By deciphering every step leading to pathogen entry, we aim to discover key interactions that will further our fundamental understanding of host-pathogen interactions and help develop more potent inhibitors of the process. Our major focus is on respiratory viruses such as Human parainfluenza virus type 3 (HPIV3) and COVID-19.